import math
import matplotlib.pyplot as plt
import torch
from IPython.display import HTML
from matplotlib.animation import FuncAnimation
from torch import tensor
from torch.distributions.multivariate_normal import MultivariateNormalClustering
Adapted from:
torch.manual_seed(42);Norms
- M.S.E. \[ \Delta x^2 + \Delta y^2 = ||v||_2 = \sqrt{|v|} = \sqrt{|v|_2} \]
- M.A.E. \[ |\Delta x | + |\Delta y | \]
- R.M.S.E. \[ \sqrt{\frac{1}{n}\Sigma_{i=0}^n (x_i - \hat{x}_i)^2} = ||v||^2_2 = |v| = |v|_2 \]
Create synthetic data
n_clusters = 4
n_samples = 256
def sample(mean):
covariance = torch.diag(tensor([5.0, 5.0], dtype=torch.float))
return MultivariateNormal(mean, covariance).sample((n_samples,))
centroids = torch.rand(n_clusters, 2) * 70 - 35
data_by_centroid = [sample(mean) for mean in centroids]
data = torch.cat(data_by_centroid)
fig, ax = plt.subplots(figsize=(4, 4))
for subset in data_by_centroid:
ax.scatter(*subset.T, alpha=0.4)
ax.scatter(*centroids.T, marker="x", color="black")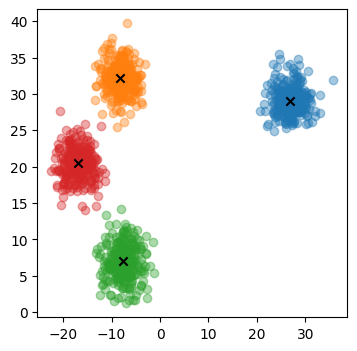
Mean Shift
Neat algorithm that has some unique advantages compared to K-means, because it does not require setting the number of clusters in advance. It does, however, require a “bandwidth” parameter and that the clusters have a Gaussian falloff.
The algorithm is as follow:
- For each point, compute the distance between that point and all other points
- Reassign each point as the weighted average for each distance with a Gaussian kernel where the variance is the “bandwidth” hyperparameter
- Repeat until convergence
The bandwidth should be about a third of the data, so in this case its about 25.
bandwidth = 2.5
def gaussian(x, mu, sig):
return (
1.0
/ (math.sqrt(2.0 * torch.pi) * sig)
* torch.exp(-torch.pow((x - mu) / sig, 2.0) / 2)
)
def gkernel(x):
return gaussian(x, mu=0, sig=bandwidth)
x = torch.linspace(0, 10, 100)
y = gkernel(x)
fig, ax = plt.subplots()
ax.plot(x, y)
ax.set(ylabel="$P.D.F.(X)$", xlabel="$X$")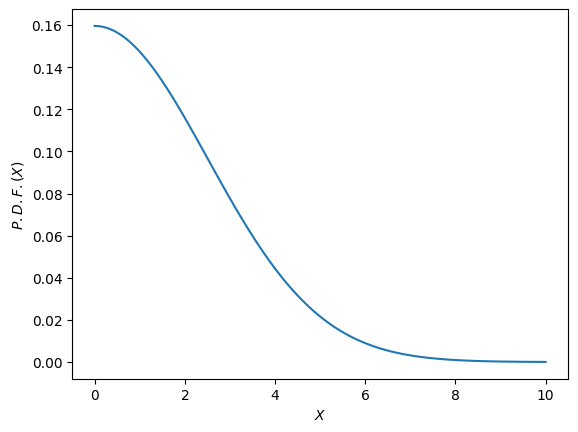
# Manipulate a copy of the data
X = data.clone()
x, *_ = X
x - Xtensor([[ 0.0000, 0.0000],
[ 4.5407, 1.0199],
[ 4.5527, 1.6327],
...,
[47.1212, 9.3284],
[50.7346, 6.5993],
[44.9273, 8.0535]])e = ((x - X) ** 2).sum(axis=1).sqrt()
etensor([ 0.0000, 4.6538, 4.8366, ..., 48.0356, 51.1620, 45.6434])weight = gkernel(e)
weighttensor([0.1596, 0.0282, 0.0246, ..., 0.0000, 0.0000, 0.0000])Now, we have the weights and the data, but we need to combine them.
weight.shape, X.shape(torch.Size([1024]), torch.Size([1024, 2]))# This doesn't work
try:
weighted_rows = weight * X
except RuntimeError as e:
print(e)The size of tensor a (1024) must match the size of tensor b (2) at non-singleton dimension 1# This does, such that the weight is copied to both target
# dimensions by broadcasting
weighted_rows = weight[:, None] * X
weighted_rowstensor([[4.9080, 4.4652],
[0.7397, 0.7607],
[0.6435, 0.6471],
...,
[-0.0000, 0.0000],
[-0.0000, 0.0000],
[-0.0000, 0.0000]])# We need to take a weighted sum, but notice that the weights
# add up to 1200%. We need to normalize.
weight.sum()tensor(12.1010)weighted_rows.sum(axis=0) / weight.sum()tensor([28.6881, 28.6938])X = data.clone()
def iteration(X):
for i, x in enumerate(X):
delta = x - X
dist = torch.einsum("ij,ij->i", delta, delta).sqrt()
weight = gkernel(dist)
weighted_rows = weight[:, None] * X
X[i, :] = weighted_rows.sum(axis=0) / weight.sum()
for _ in range(5):
iteration(X)
plt.scatter(*X.T)
plt.scatter(*centroids.T, marker="x")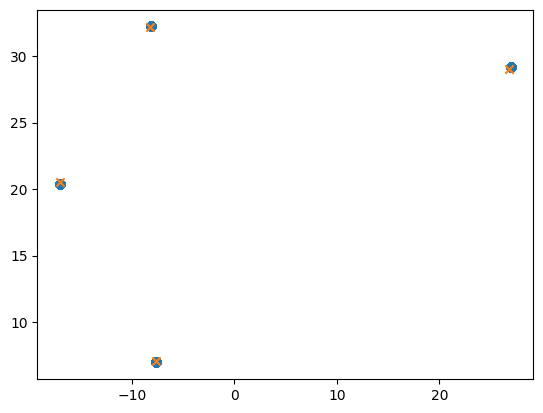
X = data.clone()
def do_one(d):
if d:
iteration(X)
ax.clear()
ax.scatter(*X.T)
ax.scatter(*centroids.T, marker="x")
fig, ax = plt.subplots()
anim = FuncAnimation(fig, do_one, frames=5, interval=500, repeat=True)
plt.close()
HTML(anim.to_jshtml())How do we GPU accelerate this?
The issue is this loop:
for i, x in enumerate(X):This is called the “kernel launching overhead”. Let’s try to batch this operation
bs = 5X = data.clone()
miniX = X[:bs]
miniXtensor([[30.7565, 27.9813],
[26.2158, 26.9614],
[26.2037, 26.3486],
[25.6908, 30.2133],
[23.2542, 28.5149]])Now, we need to compute the matrix of distances for each row, which is a \(m \times n\) matrix, where \(m\) is the bs (batch size) and \(n\) is the dataset size.
Now, we can’t just subtract these off because of the mismatched first dimension.
try:
X - miniX
except RuntimeError as e:
print(e)The size of tensor a (1024) must match the size of tensor b (5) at non-singleton dimension 0Let’s try adding some unit dimensions
X[None, :].shape, miniX[:, None].shape(torch.Size([1, 1024, 2]), torch.Size([5, 1, 2]))These dimensions are broadcast compatible because they will result in a copy of the non-batch dimension across the new-unit dimension. Let’s see what happens when we do the subtraction now.
delta = X[None, :] - miniX[:, None]
delta.shapetorch.Size([5, 1024, 2])Now, we can index into the \(row_i\) and \(row_j\) difference as desired
delta[0, 0, :], delta[0, 1, :](tensor([0., 0.]), tensor([-4.5407, -1.0199]))Now, we just need to collapse this into a \(bs \times n\) matrix of distances
dist = (delta**2).sum(axis=2).sqrt()
disttensor([[ 0.0000, 4.6538, 4.8366, ..., 48.0356, 51.1620, 45.6434],
[ 4.6538, 0.0000, 0.6129, ..., 43.3835, 46.5297, 40.9945],
[ 4.8366, 0.6129, 0.0000, ..., 43.2585, 46.4482, 40.8819],
[ 5.5356, 3.2940, 3.8986, ..., 43.6154, 46.5149, 41.1672],
[ 7.5212, 3.3443, 3.6595, ..., 40.8279, 43.8168, 38.3975]])Alternately, as an einsum:
torch.einsum("ijk,ijk->ij", delta, delta).sqrt()tensor([[ 0.0000, 4.6538, 4.8366, ..., 48.0356, 51.1620, 45.6434],
[ 4.6538, 0.0000, 0.6129, ..., 43.3835, 46.5297, 40.9945],
[ 4.8366, 0.6129, 0.0000, ..., 43.2585, 46.4482, 40.8819],
[ 5.5356, 3.2940, 3.8986, ..., 43.6154, 46.5149, 41.1672],
[ 7.5212, 3.3443, 3.6595, ..., 40.8279, 43.8168, 38.3975]])weights = gkernel(dist)
weightstensor([[0.1596, 0.0282, 0.0246, ..., 0.0000, 0.0000, 0.0000],
[0.0282, 0.1596, 0.1549, ..., 0.0000, 0.0000, 0.0000],
[0.0246, 0.1549, 0.1596, ..., 0.0000, 0.0000, 0.0000],
[0.0138, 0.0670, 0.0473, ..., 0.0000, 0.0000, 0.0000],
[0.0017, 0.0652, 0.0547, ..., 0.0000, 0.0000, 0.0000]])weights.shape, X.shape(torch.Size([5, 1024]), torch.Size([1024, 2]))norm_constant = 1 / weights.sum(axis=1)
# Note that we need to broadcast the normalization over the X and Y dimensions
reweighted = (weights @ X) * norm_constant[:, None]
reweightedtensor([[28.6881, 28.6938],
[26.6096, 28.3607],
[26.5901, 28.1598],
[26.3962, 29.4940],
[25.2566, 28.8559]])def mean_shift_batch(data, bs=512, iterations=5):
X = data.clone()
n = len(X)
for _ in range(iterations):
for i in range(0, n, bs):
batch_idxs = slice(i, min((i + bs, n)))
batch = X[batch_idxs]
delta = X[None, :] - batch[:, None]
weight = gkernel(torch.einsum("ijk,ijk->ij", delta, delta).sqrt())
norm = 1 / weight.sum(axis=1)
X[batch_idxs] = (weight @ X) * norm[:, None]
return XDEVICE = (
"mps"
if torch.backends.mps.is_available()
else "cuda"
if torch.cuda.is_available()
else "cpu"
)data_acc = data.to(DEVICE)mean_shift_batch(data_acc)tensor([[ 26.9455, 29.1479],
[ 26.9455, 29.1479],
[ 26.9455, 29.1479],
...,
[-16.9937, 20.3887],
[-16.9937, 20.3887],
[-16.9937, 20.3887]], device='mps:0')Homework
Implement DBSCAN
X = data.clone()n, _ = X.shapeeps = 2.5
min_points = 25norms = torch.pow(X[None, :] - X[:, None], 2).sum(axis=2).sqrt()
normstensor([[ 0.0000, 4.6538, 4.8366, ..., 48.0356, 51.1620, 45.6434],
[ 4.6538, 0.0000, 0.6129, ..., 43.3835, 46.5297, 40.9945],
[ 4.8366, 0.6129, 0.0000, ..., 43.2585, 46.4482, 40.8819],
...,
[48.0356, 43.3835, 43.2585, ..., 0.0000, 4.5282, 2.5374],
[51.1620, 46.5297, 46.4482, ..., 4.5282, 0.0000, 5.9866],
[45.6434, 40.9945, 40.8819, ..., 2.5374, 5.9866, 0.0000]])cluster_assignment = torch.zeros(n)
cluster_assignmenttensor([0., 0., 0., ..., 0., 0., 0.])from functools import lru_cache
from typing import Optional
class DontReassignCluster(Exception):
...
@lru_cache
def core_point_neighbords(point_idx) -> Optional:
dists = norms[point_idx, :]
neighboring_point_idxs = dists < eps
if neighboring_point_idxs.sum() > min_points:
return neighboring_point_idxs
def assign_cluster(cluster_id, point_idx):
if cluster_assignment[point_idx]:
raise DontReassignCluster
# First, assign point to cluster
cluster_assignment[point_idx] = cluster_id
# Then, if it is a core point, recursively assign the neighbors
dists = norms[point_idx, :]
neighboring_point_idxs = core_point_neighbords(point_idx)
if neighboring_point_idxs is not None:
for i, is_neighbor in enumerate(neighboring_point_idxs):
if is_neighbor:
try:
assign_cluster(cluster_id, i)
except DontReassignCluster:
continue
current_cluster_id = 1
for point_idx in range(n):
if core_point_neighbords(point_idx) is not None:
try:
assign_cluster(current_cluster_id, point_idx)
except DontReassignCluster:
continue
else:
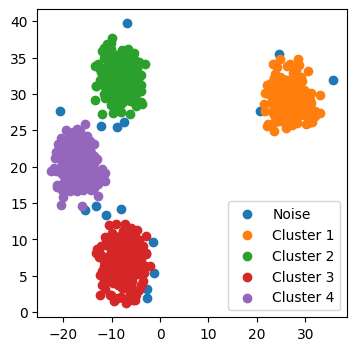
current_cluster_id += 1fig, ax = plt.subplots(figsize=(4, 4))
for cluster in cluster_assignment.unique():
ax.scatter(
*X[cluster_assignment == cluster, :].T,
label=f"Cluster {int(cluster)}" if cluster else "Noise",
)
ax.legend();